The DENV RNA genome encodes a single long open reading frame that encodes multiple viral proteins. Examining the DENV genome, the first region that we encounter is the 5′ untranslated region (5′-UTR). This is a short but highly structured sequence of between 95 to 101 nucleotides, which contains a wide variety of RNA signals, including information that regulates the fundamental processes of the virus life cycle.
Generally, the 5′-UTR is positioned upstream of the start codon and it is never translated. This region is transcribed with the coding region and is present in the mature mRNA. The 5′-UTR plays important roles in mRNA stability, mRNA localization, and translational efficiency, ultimately controlling gene expression. The ability of the 5′-UTR to perform these functions depends on its sequence and the structures it forms.
SLA and SLB structures
RNA virus genomes are compact carriers of biological information, encoding information required for replication both in their primary sequences and in the higher-order structures that are formed when the RNA genome strand folds back on itself. These 3D structures promote a compact overall architecture and contribute to viral fitness. One-third of the dengue virus RNA genome forms higher-order interactions, many in regions functionally important for replication.
The DENV 5′-UTR contains two defined RNA structures that have distinct functions during the process of viral RNA synthesis. The first domain of ∼70 nucleotides is predicted to fold into a large stem-loop A (SLA), and it is essential for viral RNA synthesis because it is recognized by the NS5 polymerase. The SLA contains three helical regions (S1, S2, and S3), a top loop, and a side stem-loop. Substitution of nucleotides at the top loop of the SLA promoter impairs RNA synthesis in vitro and viral replication in cells. Stem formation is disrupted in infectious DENV RNAs carrying mutations at each side of the stems (S1 and S2), and these mutant viruses also show impaired replication. Also, a short poly(U) tract located immediately downstream of the SLA is necessary for RNA synthesis, although it is not involved in genome circularization (see scheme below).
The second domain of the DENV 5′-UTR is predicted to form a short stem-loop (SLB) that ends in the translation initiation codon (AUG). The SLB contains a 16-nucleotide sequence known as the 5′UAR, which is complementary to a sequence located at the 3′ end of the viral genome, and that mediates long-range RNA-RNA interaction and genome cyclization.
For dengue and other flaviviruses, the circular conformation of the genome is a crucial determinant for viral replication. In our next DENV-themed article, we’ll describe how these structures in the 5′-UTR combine with other structures in the capsid region and the 3′UTR to exert their functions.
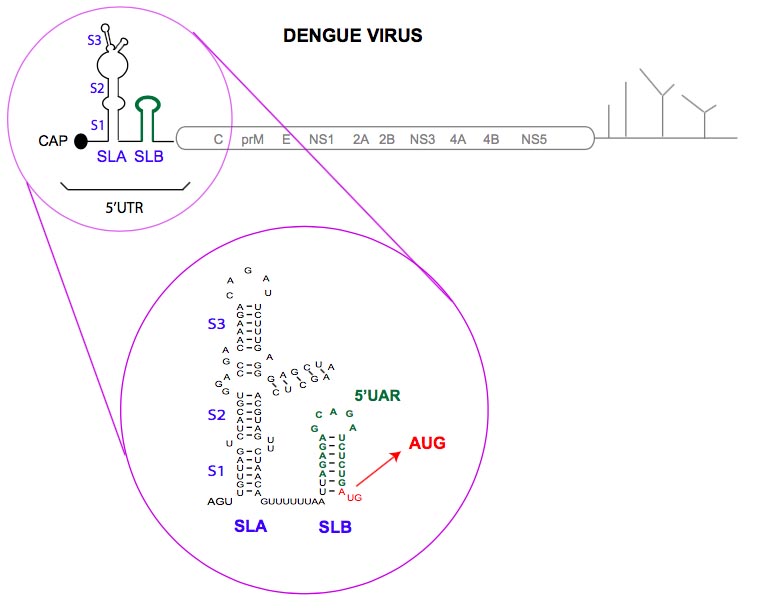
Secondary structure of DENV 5’UTR
The 5′-UTR of Dengue, a minute IRES
DENV controls its translation by a “cap-dependent” (m7GpppANNN…) mechanism but the DENV genome can also be translated through a noncanonical mechanism that appears not to require a functional cap structure (see here). Many studies have shown that DENV can also initiate protein synthesis using a cap-independent mechanism: in fact, it was demonstrated that a noncapped transcript DENV RNA can readily infect mammalian (BHK and Vero) or mosquito cells (C6/36) and produce virus at high titers. This is possible because the 5′-UTR of DENV harbors a region that, independently of a cap structure, is capable of initiating translation. The DENV2 5′-UTR, therefore, can function as a minute internal ribosomal entry site element (IRES) that induces robust translation of a downstream gene.
IRES elements are among the most mysterious RNA structures in eukaryotic molecular biology, and the origin of IRES elements during evolution is unknown. All IRESs are RNA segments that require the cooperation of cellular protein factors for function. Since single-stranded RNAs can fold and form innumerable structures, one can envise that one or more of these structures can lead to complexes between the ribosomal subunits, canonical translation factors, and possibly, cellular proteins.
The cellular proteins necessary for the IRES functions of the DENV 5′-UTRs are not yet known. A role for IRES in the 5′ UTR of DENV could have emerged during virus infection when cap-dependent translation is suppressed to inhibit the proliferation of the invading pathogen, or when DENV continues its translation through metaphase (in this stage cap-dependent translation is physiologically decreased).
Viral RNA genomes are highly dynamic molecules that form secondary and tertiary structures, which regulate multiple viral processes in the infected cell. In the next episodes, we will see how RNA plasticity in DENV is governed by local and long-range RNA-RNA interactions all determined by short nucleotide sequences distributed along its entire genome.